Part:BBa_R0085
T7 Consensus Promoter Sequence
The T7 promoter should only produce PoPS when the T7 polymerase is also being expressed.
Literature Characterization by AFCM-Egypt 2022 team
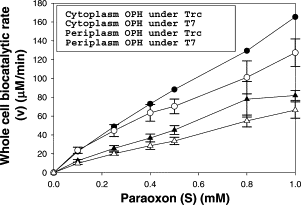
In this study, in comparison to the cytoplasmically-expressing strain, the periplasmic-secreting strain showed faster organophosphorus OPH biocatalytic reaction rates. Additionally, the bacteria under T7 promoter regulation showed faster biocatalytic reaction rates than the strains under Trc promoter regulation, irrespective of the location of OPH inside the cell. The periplasmic-secreting strain's biocatalytic rate was substantially higher with substrate concentrations than the cytoplasmic-expressing strain and showed a 2-fold better conversion rate with 1 mM Paraoxon as shown in figure 2.
Experimental Characterization by AFCM-Egypt 2022 team
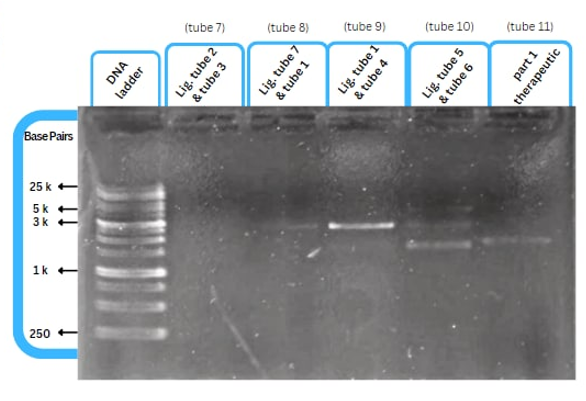
This figure shows an experimental characterization of this part as it's validated through gel electrophoresis as it is in lane 6 (the last one). The run part (ordered from IDT) included T7P - TyrR RBS - TyrR - TyrPromoter.
Exeter Team 2024
The Exeter iGEM 2024 team are designing a rapid detection system for Bovine Tuberculosis (bTB) using CRISPR-Cas detection systems. The literature suggests that bTB infection in cattle can be detected by nucleic acid biomarkers in both blood [2] and tissue samples [3]. Therefore, there was potential to develop tests looking for both DNA and RNA biomarkers in infected cattle.
For our project we needed to express both Lachnospiraceae bacterium ND2006 Cas12a and Leptotrichia wadei Cas13a enzymes at high levels, but we also wanted to control when our enzymes would be expressed. Our supervisor has previously used the IPTG inducible promoter system in the Novagen pET plasmids for over expression of proteins. This includes the T7 bacteriophage promoter in combination with the E. coli lac operator sequence and the strong RBS from T7 bacteriophage gene 10. Transcription in the pET plasmids is terminated by the transcription terminator for bacteriophage T7 RNA polymerase. There are many versions of these sequences in the Registry of Standard Biological Parts but the ones most similar to the pET system are:
• R0085- T7 promoter sequence
• BBa_K4673024- E. coli Lac operator
• BBa_K3257011- T7 gene 10 RBS
• BBa_K395601- T7 RNA polymerase terminator
For expression of our enzymes, a composite part comprising R0085, K4673024 and K3257011 with Type IIS prefix and suffix sequences was ordered as a gBlock from IDT (see BBa_K5124042). K395601 was also ordered as a gBlock from IDT with Type IIS prefix and suffix sequences. These were cloned into a medium copy plasmid (origin of replication from pBR322 [4]) carrying an ampicillin selection marker with either the coding sequence for LbCas12a (BBa_K5124000) or LwCas13a (BBa_K5124001) using Type IIS cloning.
The resulting expression plasmids were transformed into E. coli BL21(DE3) (Novagen) and protein expression induced by autoinduction media [5]. The enzymes were purified via Ni-affinity and size exclusion chromatography. Please see our Wiki for the detailed protocol (Wiki).
Both enzymes were successfully expressed and purified as verified by SDS-PAGE and Western Blot analysis (Figure 3 and 4). For further results please see BBa_K5124000 or BBa_K5124001.


Usage and Biology
This promoter is the consensus T7 promoter running from -17 to +6.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
1. Kang, D. G., Seo, S. H., Choi, S. S., & Cha, H. J. (2006). Comparison of whole cell biocatalytic reaction kinetics for recombinant Escherichia coli with periplasmic-secreting or cytoplasmic-expressing organophosphorus hydrolase. Studies in surface science and catalysis, 173-176.
2. McLoughlin KE, Correia CN, Browne JA, Magee DA, Nalpas NC, Rue-Albrecht K, et al. RNA-Seq Transcriptome Analysis of Peripheral Blood From Cattle Infected With Mycobacterium bovis Across an Experimental Time Course. Frontiers in Veterinary Science. 2021; 8:662002.
3. Taylor GM, Worth DR, Palmer S, Jahans K, Hewinson RG. Rapid detection of Mycobacterium bovis DNA in cattle lymph nodes with visible lesions using PCR. BMC Vet Res. 2007 Jun 13; 3:12.
4. Sutcliffe JG. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979; 43 Pt 1:77-90.
5. Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005 May; 41(1):207-34.
Improvement of R0085 by ZJU-CHINA 2018 Teams
The ZJU-CHINA 2018 Teams fulfilled the improvement of R0085 by inducing some mutation in the sequence. A series of promoters with increased expression strength are constructed. The result can be seen by clicking the link below.
<a href=https://parts.igem.org/Part:BBa_K2721000>BBa_K2721000</a>
<a href=https://parts.igem.org/Part:BBa_K2721001>BBa_K2721001</a>
<a href=https://parts.igem.org/Part:BBa_K2721002>BBa_K2721002</a>
<a href=https://parts.igem.org/Part:BBa_K2721003>BBa_K2721003</a>
//direction/forward
//chassis/prokaryote/ecoli
//promoter
//regulation/constitutive
//classic/regulatory/uncategorized
//chassis/bacteriophage/T7
| negative_regulators | |
| positive_regulators |